First, we plot a "correlation heatmap" using the same logic that Martin uses. In our example, let's use the Movies dataset that comes with ggplot2.
We take the 6 genre columns, and we can compute the correlation matrix for those 6 columns.
Here's what the matrix looks like:
> cor(movieGenres) # 6x6 cor matrix
Action Animation Comedy Drama
Action 1.000000000 -0.05443315 -0.08288728 0.007760094
Animation -0.054433153 1.00000000 0.17967294 -0.179155441
Comedy -0.082887284 0.17967294 1.00000000 -0.255784957
Drama 0.007760094 -0.17915544 -0.25578496 1.000000000
Documentary -0.069487718 -0.05204238 -0.14083580 -0.173443622
Romance -0.023355368 -0.06637362 0.10986485 0.103545195
Documentary Romance
Action -0.06948772 -0.02335537
Animation -0.05204238 -0.06637362
Comedy -0.14083580 0.10986485
Drama -0.17344362 0.10354520
Documentary 1.00000000 -0.07157792
Romance -0.07157792 1.00000000
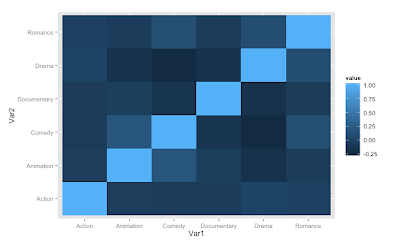
When we plot with the default colors we get:
It is difficult to see the details in the tiles. Now, if you want to better control the colors, you can use the handy colorRampPalette() function and combine that with scale_fill_gradient2.
Let's say that we want "red" colors for negative correlations and "green" for positives.
(We can gray out the 1 along the diagonal.)
Doing this produces:
If there are values close to 1 or to -1, those will pop out visually. Values close to 0 are a lot more muted.
Hope that helps someone.
References: Using R: Correlation Heatmap with ggplot2